Dynamical Systems & Modeling
Research
During my graduate research with Dr John Tyson, I built mathematical models to understand dynamical aspects:
-
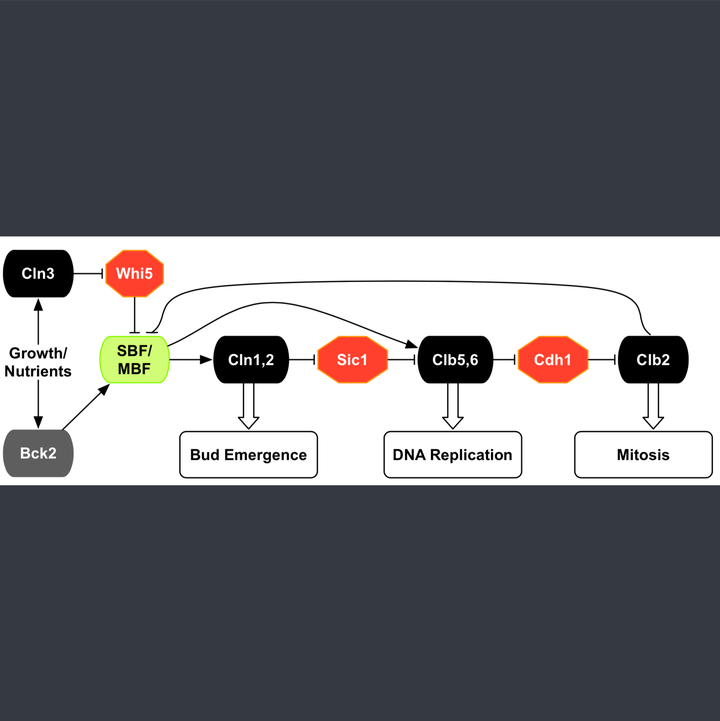
Modeling the START transition in the budding yeast cell cycle:
- Built a detailed mathematical model (~100 ODEs, ~150 parameters) for the START transition in yeast and integrated it with our published model of the whole cell cycle.
- Model addresses outstanding issues related to the precise mechanism and timing of transcriptional, post-translational and localization events, as well as size control under varying growth conditions.
- Model consistent with ~200 experimental mutant phenotypes pertaining to the START transition and rest of the cell cycle.
- Built a basic model for the nutritional effect of size control in budding yeast cells.
- This mechanism has been incorporated into the existing model of the yeast cell cycle to explain an initial set of START mutants.
-
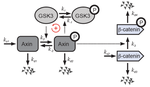
Modeling bistability in the canonical Wnt pathway:
- Built a simplified model based upon the core module of the Wnt canonical pathway, and incorporated additional key regulatory interactions.
- Model shows that the Wnt signaling pathway can display bistability, in agreement with preliminary experimental results.
Related publications
- Ravi J#, Samart K, Zwolak JW, Tyson JJ#. Modeling the START transition in the budding yeast cell cycle. Model Submitted. #Co-corresponding authors.
- Cantoria MJ*, Alizadeh E*, Ravi J, Bunnag N, Kettenbach N, Ahmed Y, Paek AL, Tyson JJ, Doubrovinski K#, Lee E#, Thorne CA#. Feedback in the β-catenin destruction complex imparts bistability and cellular memory. 2022 bioRxiv
Students
- Kewalin Samart
Collaborators
Virginia Tech
- John Tyson
Vanderbilt University
- Ethan Lee
- Curtis Thorne (now at University of Arizona)