Abstract
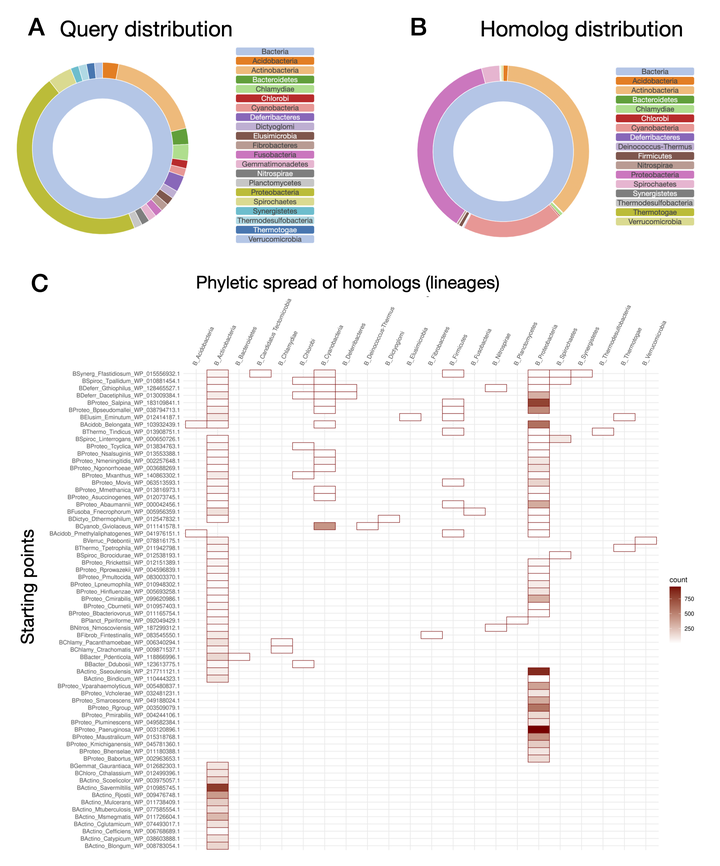
A fundamental requirement for life is the replication of an organism’s DNA. Studies in Escherichia coli and Bacillus subtilis have set the paradigm for DNA replication in bacteria. During replication initiation in E. coli and B. subtilis, the replicative helicase is loaded onto the DNA at the origin of replication by an ATPase helicase loader. However, most bacteria do not encode homologs to the helicase loaders in E. coli and B. subtilis. Recent work has identified the DciA protein as a predicted helicase operator that may perform a function analogous to the helicase loaders in E. coli and B. subtilis. DciA proteins, which are defined by the presence of a DUF721 domain (termed the DciA domain herein), are conserved in most bacteria but have only been studied in mycobacteria and gammaproteobacteria (Pseudomonas aeruginosa and Vibrio cholerae). Sequences outside the DciA domain in Mycobacterium tuberculosis DciA are essential for protein function but are not conserved in the P. aeruginosa and V. cholerae homologs, raising questions regarding the conservation and evolution of DciA proteins across bacterial phyla. To comprehensively define the DciA protein family, we took a computational evolutionary approach and analyzed the domain architectures and sequence properties of DciA domain-containing proteins across the tree of life. These analyses identified lineage-specific domain architectures among DciA homologs, as well as broadly conserved sequence-structural motifs. The diversity of DciA proteins represents the evolution of helicase operation in bacterial DNA replication and highlights the need for phylum-specific analyses of this fundamental biological process.
HCB and JTB are co-primary authors. JR and CLS are co-corresponding authors.