Computational evolutionary approaches
Summary
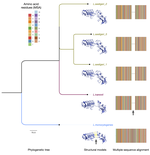
We are developing an integrated computational evolutionary framework that will bring together: 1) molecular evolution and phylogeny; and 2) comparative genomics/pangenomics to study any pathogen-group or protein family, and discover distinctive protein/genomic features relevant to biological comparisons of interest (such as identifying diagnostic or candidate vaccine targets in emerging, poorly characterized pathogens).
Team
Students
Current
- Joe Burke
- Elliot Majlessi
- Vignesh Sridhar
- Ethan Wolfe
Alumni
- Samuel Chen
- Lo Sosinski
- Karn Jongnarangsin
- Lauryn Brooks
Collaborators
- Neal Hammer, MSU (Staphylococci)
- Srinand Sreevatsan, Robert Abramovitch, and Andrew Olive (MSU), Christina Stallings, WUSTL (Mycobacteria)
- Antonella Fioravanti, VUB Brussels (B. anthracis)
- Chris Waters, MSU (Vibrio)
- Jonathan Hardy, MSU (Listeria)
- Stephanie Shames, KSU (Legionella)
- Priya Shah, UCD (Flaviviruses)
- Sean Crosson, MSU (Brucella, Caulobacter)
Janani Ravi
Assistant Professor
My research interests include computational pathogenomics and host-directed drug-repurposing.
Related
- Bacterial Stress Response
- Phage-shock-protein (Psp) Envelope Stress Response: Evolutionary History & Discovery of Novel Players
- Computational evolutionary approaches for understanding pathogen proteins and genomes
- Phylogenetics & Comparative Genomics | MMG801
- Computational evolutionary approaches to characterize bacterial proteins
Publications
The glutathione import system satisfies the Staphylococcus aureus nutrient sulfur requirement and promotes interspecies competition
DciA Helicase Operators Exhibit Diversity across Bacterial Phyla
Novel Internalin P homologs in Listeria ivanovii londoniensis and Listeria seeligeri
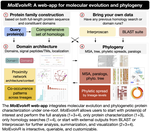
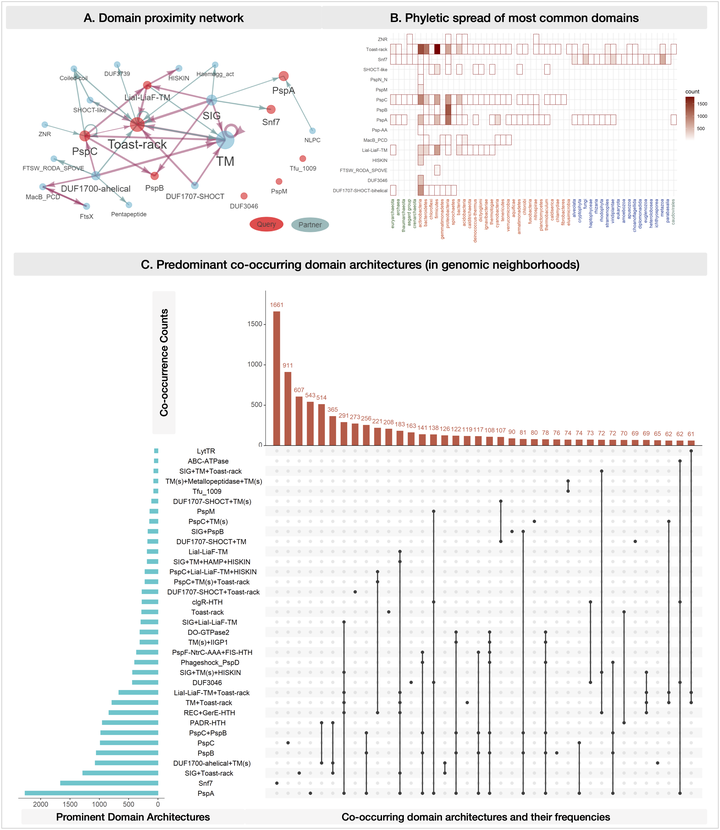
MolEvolvR: a web-app for characterizing proteins using molecular evolution and phylogeny
Discovery of a predominant and distinct lineage of Mycobacterium tuberculosis in Brazilian indigenous population
Talks
MolEvolvR BTS: When and how to build a web-app and software package?
We recently developed a web application, MolEvolvR, to characterize proteins using molecular evolution and phylogeny. This session will …
Cross-database integration using evolution and machine learning to identify multiscale molecular building blocks for antibiotic resistance
Computational approaches to study molecular pathogenesis and intervention of infectious diseases
MolEvolvR: A webapp for characterizing proteins using molecular evolution and phylogeny
MolEvolvR: Web-app and R-package for characterizing proteins using molecular evolution and phylogeny
Using a computational molecular evolution and phylogeny to study pathogenic proteins
MolEvolvR: a web-app for molecular evolution and phylogeny
We will discuss the alpha version of their new easy-to-use interactive webapp: MolEvolvR, for characterizing proteins using molecular …