MolEvolvR: a web-app for characterizing proteins using molecular evolution and phylogeny
Abstract
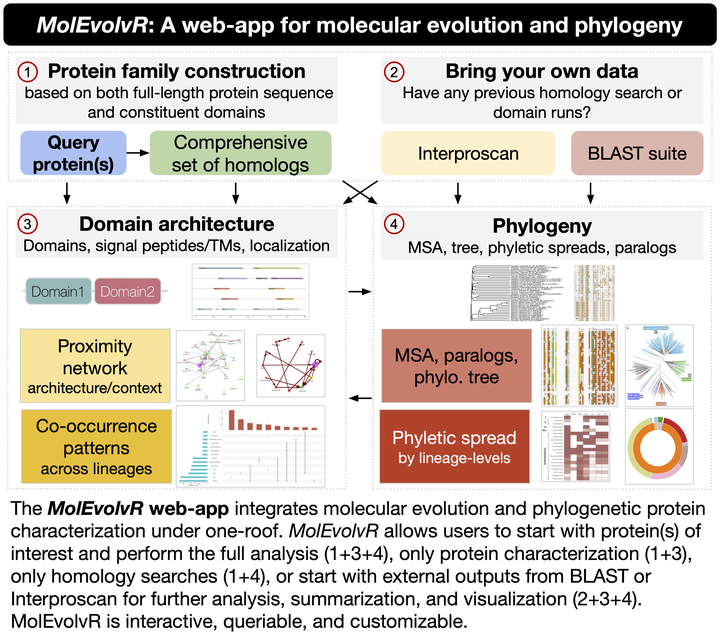
Studying proteins through the lens of evolution can help identify conserved features and lineage-specific variants, and potentially, their functions. MolEvolvR ( http://jravilab.org/molevolvr) is a web-app that enables researchers to run a general-purpose computational workflow for characterizing the molecular evolution and phylogeny of their proteins of interest. The web-app accepts input in multiple formats: protein/domain sequences, homologous proteins, or domain scans. MolEvolvR returns detailed homolog data, along with dynamic graphical summaries, e.g., MSA, phylogenetic trees, domain architectures, domain proximity networks, phyletic spreads, and co-occurrence patterns across lineages. Thus, MolEvolvR provides a powerful, easy-to-use interface for computational protein characterization.
JTB, SZC, LMS are co-primary authors who contributed equally; they are listed alphabetically.