Computational evolutionary approaches
Summary
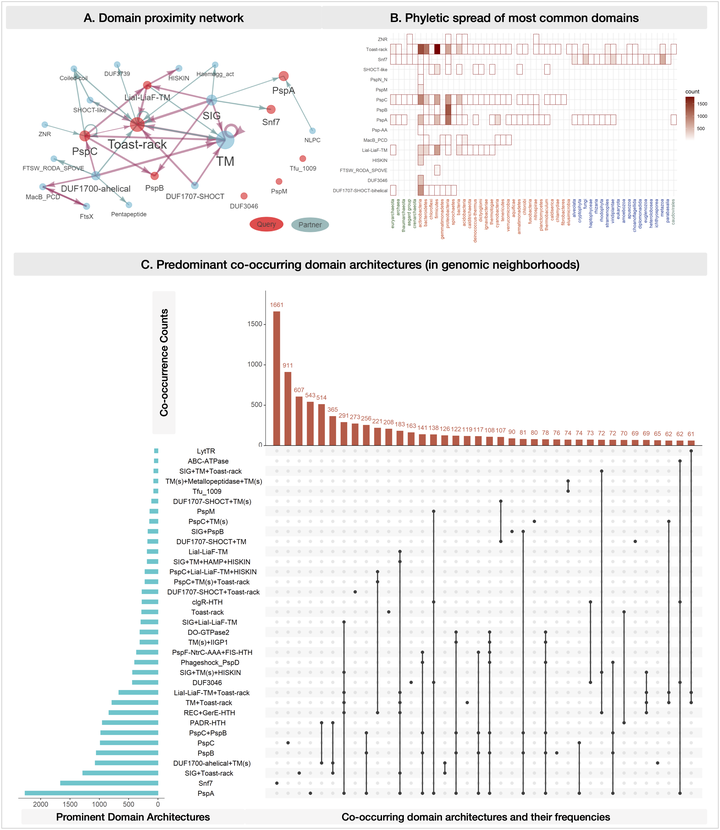
We are developing an integrated computational evolutionary framework that will bring together: 1) molecular evolution and phylogeny; and 2) comparative genomics/pangenomics to study any pathogen-group or protein family, and discover distinctive protein/genomic features relevant to biological comparisons of interest (such as identifying diagnostic or candidate vaccine targets in emerging, poorly characterized pathogens).
Team
Students
Current
- Joe Burke
- Elliot Majlessi
- Vignesh Sridhar
- Ethan Wolfe
Alumni
- Samuel Chen
- Lo Sosinski
- Karn Jongnarangsin
- Lauryn Brooks
Collaborators
- Neal Hammer, MSU (Staphylococci)
- Srinand Sreevatsan, Robert Abramovitch, and Andrew Olive (MSU), Christina Stallings, WUSTL (Mycobacteria)
- Antonella Fioravanti, VUB Brussels (B. anthracis)
- Chris Waters, MSU (Vibrio)
- Jonathan Hardy, MSU (Listeria)
- Stephanie Shames, KSU (Legionella)
- Priya Shah, UCD (Flaviviruses)
- Sean Crosson, MSU (Brucella, Caulobacter)